Development and Characterization of EST-SSR Markers from NCBI and cDNA Library in Cultivated Peanut (Arachis hypogaea L.) 
 Author
Author  Correspondence author
Correspondence author
Legume Genomics and Genetics, 2010, Vol. 1, No. 6 doi: 10.5376/lgg.2010.01.0006
Received: 15 Jun., 2010 Accepted: 06 Aug., 2010 Published: 12 Nov., 2010
Wang et al., 2009, Development and Characterization of EST-SSR Markers from NCBI and cDNA Library in Cultivated Peanut (Arachis hypogaea L.), Molecular Plant Breeding, 7(4): 806-810 (doi: 10.3969/mpb.007.000806)
86 132 ESTs downloaded from GenBank in NCBI and 12 501 ESTs from cDNA library constructed by high-oil linoleic acid accession E12 were analysed. After the preprocession, there were 18 051 singletons and 9 972 contigs in the GenBank of NCBI and cDNA library. Totally 3 104 SSR locis had been screened by MISA software, accounting for 11.08% for these non-redundant ESTs. All SSR locis are divided into di-nucleotide, thi-nucleotide, tetra-nucleotide, penta-nucleotide, hexa-nucleotide and multi-nucleotide etc., and thi-nucleotide motif is the most motifs and the frequency was 43.0% and 56.8% in NCBI and cDNA libraray, respectively. The number of di- and penta-nucleotide motifs were second and third in all motifs. And the hexa-nucleotide was the least motif both in NCBI and cDNA library. In all repeat motifs nucleotide, AG/TC was the most motifs and accounted for 8.65% and 13.42% in NCBI and cDNA library respectively. Among the tri-nucleotide repeats, CTT/GAA was the most frequent motif, accounting for 6.7% and 13.42%, respectively. The repeat unit number of SSR locis is between 4 and 51.
Peanut, or groundnut (Arachis hypogaea L., 2n=4×=40), as a source of oil and protein, is the second-most important grain legume crop after soybean in most tropical and subtropical areas of the world (Dwivedi et al., 2003). The seed is comprised of around 50% oil, of which approximately 80% consist of oleic acid (36%~67%) and linoleic acids (15%~43%) (Moore and Knauft, 1989). Additionally, the largest use is for oil, with the meal being used as a high-protein dietary supplement for human and animal consumption. In China and other countries, the peanut seed oil is used mainly in the cooking. Meanwhile, peanut may be used for fodder, and the shells used for fuel or livestock feed (Savage and Keenan, 1994).
Cultivated peanut exhibits a considerable amount of variability for various morphological, physiological, and agronomic traits. However, the genetic diversity observed is much lower in DNA level by RAPD (random amplified polymorphic DNA), AFLP (amplified fragment length polymorphisms), RFLP (restriction fragment length polymorphisms) (Kochert et al., 1996; Hilu et al., 1995; Herselman, 2003). The low level of variation in cultivated peanut has been attributed to the barriers to gene flow from related diploid species to domesticated peanut as a consequence of the polyploidization event (Young et al., 1996). And the reason that few elite breeding lines and litter exotic germplasm are used in the breeding programs is other cause of the narrow genetic base.
Simple sequence repeat (SSR) markers are microsatellite loci that can be amplified by polymerase chain reaction (PCR) using primers designed for unique flanking sequences. Polymorphism is based on variation in the number of repeats in different genotypes owing to polymerase slippage and point mutations (Kruglyak et al., 1998). SSR markers are (i) highly informative, (ii) locus-specific and frequently show co-dominant inheritance, (iii) adaptable to high-throughput genotyping, and (iv) simple to maintain and distribute. In recent years, significant efforts have been made to develop the SSR markers in groundnut and more than 800 SSR markers have been gained (Hopkins et al., 1999; He et al., 2003; Ferguson et al., 2004; Cuc et al., 2008). SSR markers include genomic SSR markers and EST-derived SSR markers. Genomic SSR markers have some disadvantages. Firstly, genomic SSR markers are derived from genomic BAC library, most of which are come from the intergenic regions with no gene function. Secondly, the procedures for developing those markers are difficult, complex, high-cost. At the same time, large-scale sequencing projects have produced a large amount of single-pass sequences of complementary DNAs (cDNAs), more and more EST sequences have been developed from different plant species (http://www.ncbi.nlm.nih.gov/). ESTs contain SSR sequences in both coding and noncoding regions (Temnykh et al., 2001), and SSRs have been successfully developed from ESTs in many species (Thiel et al., 2003; Gao et al., 2004;Nicot et al., 2004; Serapion et al., 2004;Perez et al., 2005; Wang et al., 2005).
Recently, more than 80 000 ESTs are now available for peanut in GenBank of NCBI, and the number of these ESTs is increasing year by year. But development of EST-SSR markers are lagged behind compared of other species. In this research, totally 86 132 peanut ESTs from NCBI and 12 501 ESTs in cDNA library derived from E12 which comes from a Chinese landrace contains high oleic acid are analyzed. Here we report the successful development and characterization of EST-SSR markers in peanuts. These markers can enrich the molecular biological resources in peanut.
1 Results
1.1 EST sequences screening
86 132 ESTs of the EST database in GenBank were found and 12 501 ESTs from cDNA library of E12 were obtained. All the 98 633 EST sequences from NCBI and cDNA library were used for searching singletons and contigs (Table 1). In the Genbank of NCBI, there were 14 141 singletons and 9 892 contigs. And in cDNA library derived from E12, the number of singletons and contigs was 3,910 and 80 respectively. After screening EST-SSRs using MISA software in all singletons and contigs from NCBI and cDNA library, 2 463 (9.52%) EST-SSRs in NCBI and 641 (16.4%) EST-SSRs in cDNA library were found (Table 1). In NCBI database 2,443 ESTs contained one EST-SSR, and 20 ESTs had more than two EST-SSRs. In cDNA library, 641 ESTs contained one SSR loci, 82 ESTs have two EST-SSRs and four ESTs had three EST-SSRs (Table 1).
|
Table 1 EST and EST-SSR distribution in NCBI and cDNA library |
1.2 Distribution and frequency of EST-SSRs
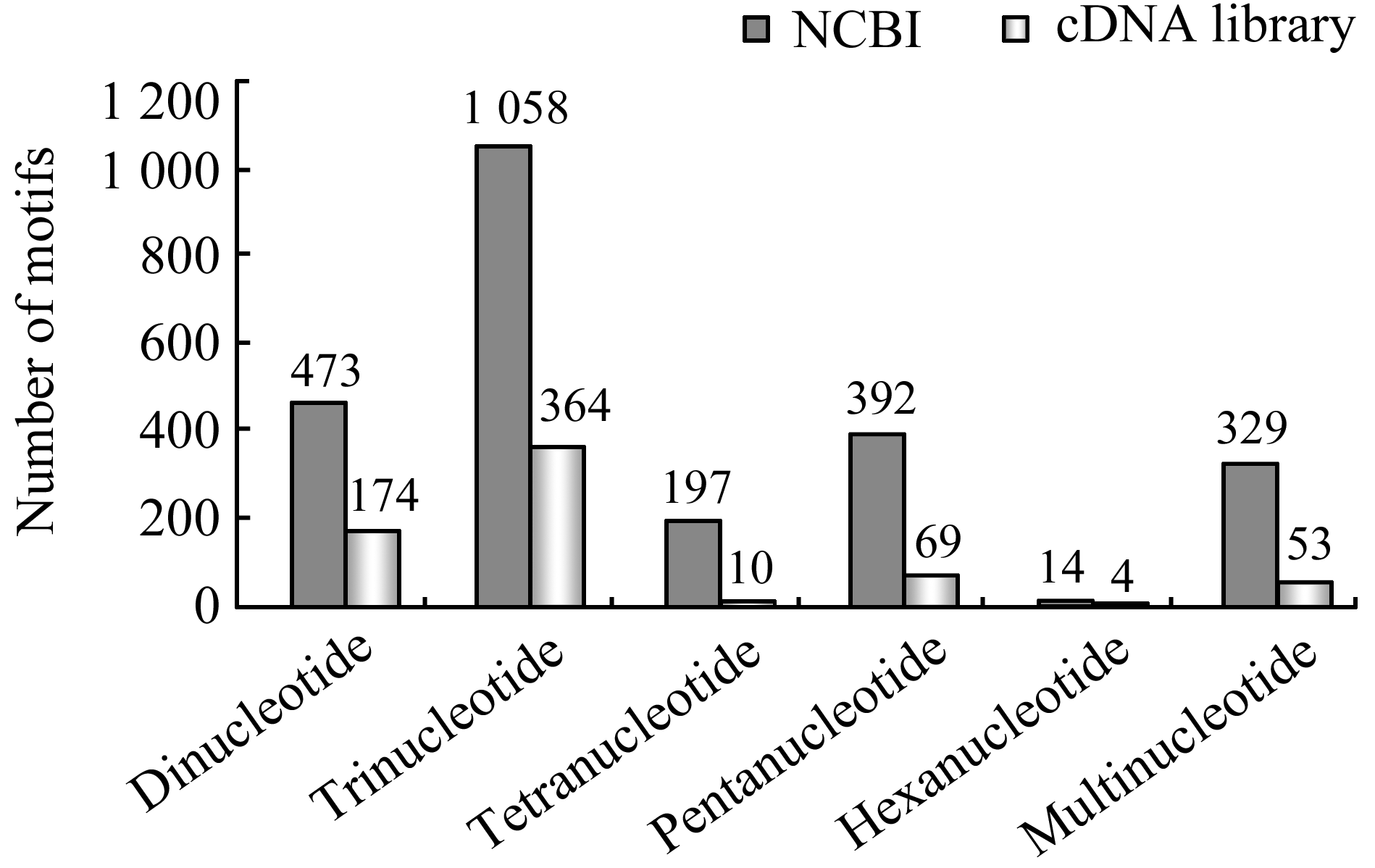
All these EST-SSRs could be divided into six kind motifs, such as di-nucleotide, thi-nucleotide, tetra-nucleotide, penta-nucleotide, hexa-nucleotide and multi-nucleotide etc., The tri-nucleotide motif was the most frequent motif in both NCBI and cDNA library. And the frequency of tri-nucleotide was 43.0% and 56.8% respectively. The number of di- and penta- nucleotide motifs was second and third in all motifs. And the hexa-nucleotide was the least motif in NCBI and cDNA library (Figure 1). The number of multi-nucleotide motif was 329 and 53, accounting for 13.4% and 8.3%, respectively.
|
Figure 1 Comparison of frequency distribution of different types of SSR motif between NCBI and cDNA library |
In terms of nucleotide repeat motifs, the AG/TC was the most motifs in all repeat types nucleotide in NCBI and cDNA library, and accounted for 8.65% and 13.42%, respectively. The following di-nucleotide repeat motif was AT/TC and CT/GA motifs. Among the tri-nucleotide repeats, CTT/GAA was the most frequent motif, accounting for 6.7% and 13.42%, respectively. And this type motif repeat is almost six and four times than the other tri-nucleotide repeats in NCBI and cDNA library respectively. The number of tetra-, penta- and hexa-nucleotide motifs is less than 2% in both NCBI and cDNA library. The AAAT/TTTA motif accounted for 1.26% in NCBI but there was no this type motif in cDNA library (Table 2).
|
Table 2 The most repeat motif of SSR loci in every type nucleotide repeat |
The maximum repeat unit number of di-nucleotide repeat motifs of AG/TC and CT/GA were 25 and 51 units in NCBI, respectively. And the numbers were 21 and 25 unit in cDNA library. In fact, in some studies, the markers developed for longer repeat motifs were found more informative for detection of polymorphism in cultivated groundnut germplasm (Moretzsohn et al. 2005).
2 Discussion
Peanut is one of important crops for both direct human food and oil production in the world. One of the major factors influencing peanut oil quality is the composition of polyunsaturated fatty acid. The linoleic acid is one kind of polyunsaturated fatty acid, and its acyl residues are susceptible to oxidation, which adversely impacts on oil stability and increases development of off-flavors commonly associated with rancidity in stored oil (Patel et al., 2004). So validating the mechanism of the polyunsaturated fatty acid synthesis and metabolism is the central goal to increase the peanut quality. In the recent research, we made use of high-oil linoleic acid accession E12 to construct the peanut cDNA library. This library contained 12 501 ESTs and 4 074 Unigene, which took part in many biological processes, such as transporting and metabolizing amino acid and carbohydrate, energy metabolism process, transcription, protein translation and modification et al., And 641 SSR loci had been screened in this library, of which 624 ESTs had been designed EST-SSR markers. The AG/TC, CT/GA and CTT/GAA repeat motifs are the most SSR motifs in all nucleotide repeat motifs.
ESTs are currently the most widely sequenced nucleotide commodity from plant genomes in terms of number of sequences and total nucleotide count. During the past few years a great deal of attention has been directed towards discovering and characterizing the range of protein-coding genes existing within the genome of plant species with large genomes. The larger size of the peanut genome is a result of polyploidy and the presence of regions with repeat motifs, both of which make it difficult to sequence the complete genome. One possible method that could be used to investigate genome coding regions is cDNA sequencing, which may be considered to be an alternative to the complete sequencing of the genome in those plants with large genomes. The availability of ESTs in public databases provides the opportunity to identify SSRs and to develop molecular markers. Consequently, the large deal of peanut EST-SSRs available developed from public databases is an important research resource which can be used to analyze the functional portion of the genomes. In the present study, there were 86 132 ESTs downloaded from Genbank in NCBI. 14 141 contigs and 9 892 singletons were obtained, these sequences contained 2 463 SSR loci and 1 943 EST-SSRs are developed. The type and frequency are as same as cDNA library.
In general, molecular markers, and microsatellites or simple sequence repeats (SSRs) in particular have proven very useful for crop improvement in many species (Gupta and Varshney, 2000). However, breeding applications using molecular markers in groundnut,,which has been limited by the low level of the genetic variation in this species. This low level of genetic variation in cultivated groundnut is attributed to its origin from a single polyploidization event that occurred relatively recently on an evolutionary time scale (Young et al., 1996). However, additional contributing factors to the low levels of molecular polymorphism observed to date could be the marker techniques used and the amount of diversity of samples tested (Singh et al., 1998). In recent years, significant efforts have been made to develop the EST-SSR markers in groundnut (Ferguson et al., 2004; Moretzsohn et al., 2005; Mace et al., 2008). They should be valuable in genome mapping and population studies. The EST-SSRs have two major advantages over genomic SSRs. First, as EST-SSRs are part of or adjacent to functional genes, they can be used for the mapping and functional analysis of candidate genes. Second, because ESTs are more conserved than average genomic sequences, EST-SSRs may be more stable and transferable across species. The successful development of EST-SSR in the cultivated peanut should encourage similar efforts in other Arachis for which a large number of ESTs are available.
3 Materials and methods
3.1 Identification of SSR-Containing ESTs
ESTs of the peanut were downloaded from the EST database of NCBI GenBank (http://www.ncbi.nlm.nih.gov/dbEST) in April 2009. And EST sequences of cDNA libraries derived from leaf tissues of high-oil acid accession E12 were obtained previously (data not shown). All sequences were downloaded or transformed to a text file in FASTA format.
3.2 ESTs sequences splicing
All EST sequences were spliced by CAP3 software (http://genome.cs.mtu.edu/sas.html). These ESTs are divided into singletons which are not spliced with other ESTs and contigs which are related to one another by overlap of their sequences.
3.3 EST-SSR screening
These singletons and contigs were screened for SSRs using the MISA software (MicroSAtellite, http://pgrc.ipk-gatersleben.de/misa/). For this study, the criteria for SSRs were set as sequences having at least eight repeats of dinucleotide and five repeats for all other repeats (tri-, tetra-, penta-, and hexa-nucleotide).
3.4 Design Primers
All SSR-containing ESTs were individually inspected for suitability for primer design. SSR-containing ESTs that contain sufficient flanking sequences of good quality (no unknown bases) were selected for primer design. Primers were designed using the PRIMER 3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), with an optimal annealing temperature of 60℃ and a fragment size between 100 bp and 300 bp. A GC clamp was added at the 3’ primer end when possible.
Acknowledgements
This research was supported by Modern Agro-industry Technology Research System (nycytx-19), National High-Tech Research and Development Plan of China (2006AA10A114; 2007AA10Z189) and National Project of Scientific and Technical Supporting Program (2008BAD97B04).
References
Ferguson M.E., Burow M.D., Schulze S.R., Bramel P.J., Paterson A.H., Kresovich S., and Mitchell S., 2004, Microsatellite identification and characterization in peanut (A. hypogaea L.). Theor. Appl. Genet. 108(6): 1064-1070 doi:10.1007/s00122-003-1535-2
Gao L.F., Jing R.L., Huo N.X., Li Y., Li X.P., Zhou R.H., Chang X.P., Tang J.F., Ma Z.Y., and Jia J.Z., 2004, One hundred and one new microsatellite loci derived from ESTs (EST-SSRs) in bread wheat, Theor. Appl. Genet., 108(7): 1392-1400 doi:10.1007/s00122-003-1554-z
Gupta P.K., and Varshney R.K., 2000, The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat, Euphytica, 113(3): 163-185 doi:10.1023/A:1003910819967
Mace E.S., Varshney R.K., Mahalakshmi V., Seetha K., Gafoor A., Leeladevi Y., and Crouch J.H., 2008, In silico development of simple sequence repeat markers within the aeschynomenoid/dalbergoid and genistoid clades of the Leguminosae family and their transferability to Arachis hypogaea, groundnut, Plant Sci., 174(1): 51-60 doi:10.1016/j.plantsci.2007.09.014
Moretzsohn M.C., Leoi L., Proite K., Guimaraes P.M., Leal-Bertioli S.C.M., Gimanes M.A., Martin W.S., Valls J.F.M., Grattapaglia D., and Bertioli D., 2005, A microsatellite-based, gene-rich linkage map for the AA genome of Arachis (Fabaceae), Theor. Appl. Genet., 111: 1060-1071 doi:10.1007/s00122-005-0028-x
Nicot N., Chiquet V., Gandon B., Amilhat L., Legeai F., Leroy P., Bernard M., and Sourdille P., 2004. Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs), Theor. Appl. Genet., 109(4): 800-805 doi:10.1007/s00122-004-1685-x
Patel M., Jung S., Moore K., Powell G., Ainsworth C., and Abbott A., 2004. High-oleate peanut mutants result from a MITE insertion into the FAD2 gene, Theor. Appl. Genet., 108(8): 1492-1502 doi:10.1007/s00122-004-1590-3
Perez F., Ortiz J., Zhinaula M., Gonzabay C., Calderon J., and Volckaert F.A., 2005, Development of EST-SSR markers by data mining in three species of shrimp: Litopenaeus vannamei, Litopenaeus stylirostris, and Trachypenaeus birdy, Mar. Biotechnol., 7(5): 554-569 doi:10.1007/s10126-004-5099-1
Serapion J., Kucuktas H., Feng J.A., and Liu Z.J., 2004. Bioinformatic mining of type I microsatellites from expressed sequence tags of channel catfish (Ictalurus punctatus), Mar. Biotechnol., 6(4): 364-377 doi:10.1007/s10126-003-0039-z
Singh A.K., Smartt J., Simpson C.E., and Raina S.N., 1998, Genetic variation vis-a-vis molecular polymorphism in groundnut, Arachis hypogaea L., Genet. Resour. Crop Evol., 45(2): 119-126 doi:10.1023/A:1008646422730
Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S., and McCouch S., 2001, Computational and experimental analysis of microsatellite in rice (Oryza sativa L.): frequency, length variation, transposon association, and genetic marker potential, Genome Research, 11(8): 1441-1452 doi:10.1101/gr.184001
Thiel T., Michalek W., Varshney R.K., and Graner A., 2003, Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.), Theor. Appl. Genet., 106(3): 411-422
Wang H.X., Li F.H., and Xiang J.H., 2005. Polymorphic EST-SSR markers and their mode of inheritance in Fenneropenaeus chinensis, Aquaculture, 249(1-4): 107-114 doi:10.1016/j.aquaculture.2005.03.041
Young N.D., Weeden N.F., and Kochert G., 1996, Genome mapping in legumes (Fam. Fabaceae), In: Paterson A.H.(ed.), Genome Mapping in Plants, Landes Co., Austin, USA, pp.211-227
. PDF(107KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jinyan Wang
. Lijuan Pan
. Qingli Yang
. Shanlin Yu
Related articles
. EST-SSR
. Peanut
. Development
. Characterization
Tools
. Email to a friend
. Post a comment
.png)
.png)


